UDE researchers use DNA origami method
New ways for artificial nanofactories
- von Jennifer Meina
- 29.07.2024
Artificial nanofactories are tiny workshops made from the human body's own molecules that are precisely designed and constructed according to a blueprint. In the future, they could help to better recognise disease markers or environmental toxins, for example, or serve as highly specific catalysts for energy conversion and storage. Researchers at the University of Duisburg-Essen have developed a model that regulates the unfolding and degradation of proteins using compartmentalisation strategies – which could open up new ways of developing artificial nanofactories. Their results have now been published in Nature Nanotechnology.
In cells, chemical reactions are organised in compartmentalised enzyme systems - in other words, reactions take place in spatially limited areas, so-called compartments. This creates specific conditions for each reaction and improves the efficiency and control of biochemical processes. Inspired by these natural systems, the research team at the University of Duisburg-Essen (UDE), led by Prof. Dr Barbara Saccà from the Center for Medical Biotechnology (ZMB) in collaboration with the laboratory of Prof. Dr Hemmo Meyer, has built an artificial structure that is divided into different compartments and mediates specific reactions. To create this, they use the DNA origami method - a technique in which DNA molecules are folded like tiny building blocks to form very small structures. "As the interactions between the DNA components are precisely known, we can accurately design the structure into which the strand folds - down to the nanometre," explains the chemist.
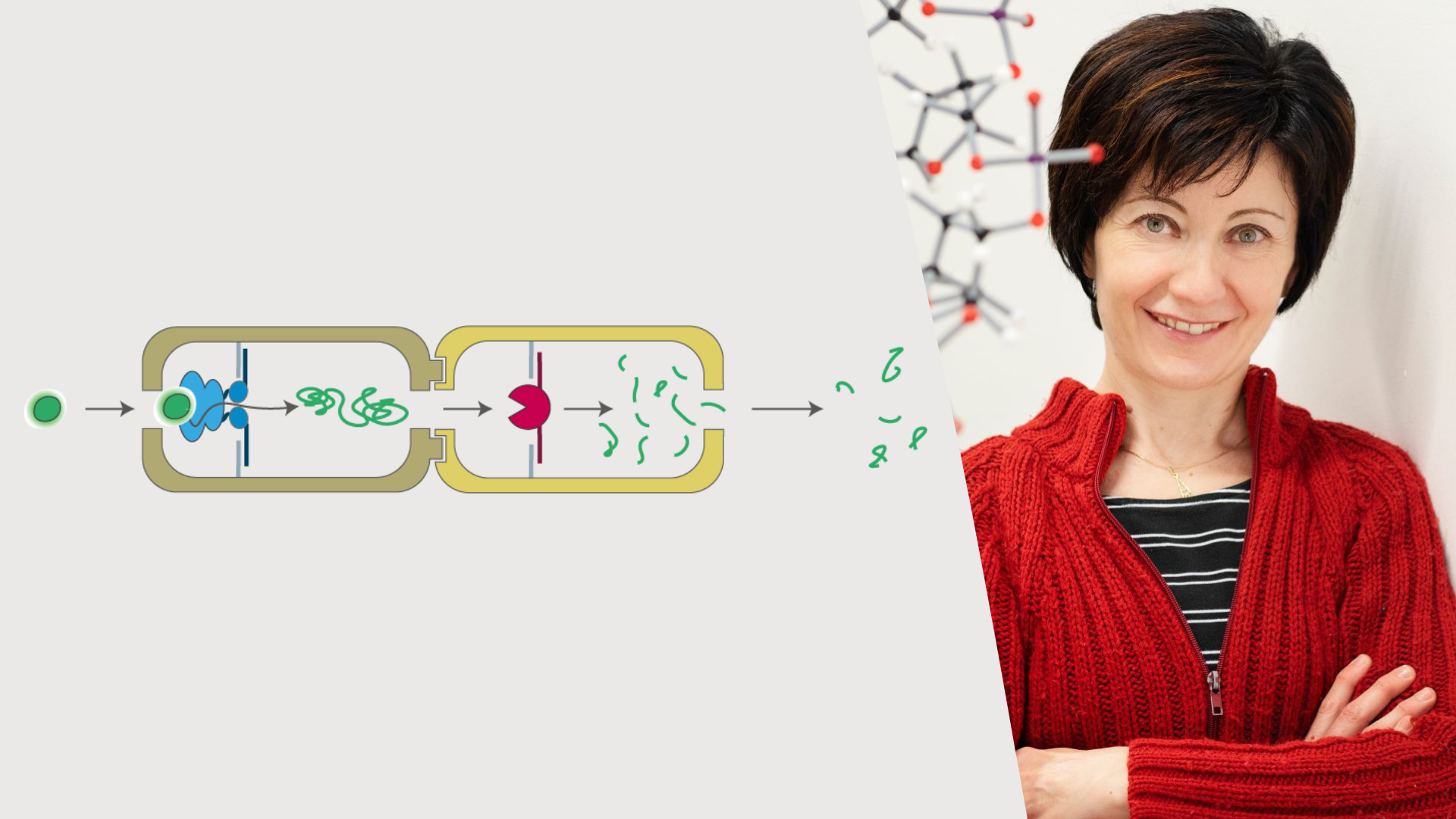
In the UDE scientists' model, the artificial nanofactory consists of two such structures, each of which encloses a cavity. The protein unfolding machine p97 is firmly anchored in the first part, which - as its name suggests - unfolds proteins. In its now elongated form, the protein is then transported to the second area in a controlled manner. A protease is bound there, which breaks down the protein strand into small fragments. The spatial connection of the two sub-processes makes the reaction significantly more efficient: the reaction speed increases tenfold and undesired side reactions are reduced almost by a factor of six compared to two separate reactions, which are also not spatially separated.
"Our findings demonstrate the potential of DNA nanotechnology for programming modular and compartmentalized enzyme systems and open up new avenues for synthetic biology and biocatalytic applications," says Saccà. In the future, the professor of bionanotechnology aims to further improve origami factories: for example (1) by coating the origami with organic or inorganic materials that mimic the concept of a semi-permeable membrane or (2) by allowing access to the reaction chamber at only one point, thus better protecting the processes inside against external influences. "Advances of this kind could lead to the development of miniature laboratories with capabilities beyond those of natural systems."
In picture:
Left: The substrate (green) is recognized by the unfolding machine p97 (blue) oriented in a unique and well-defined direction within a first nanocompartment. The substrate is then unfolded and translocated to a second compartment, hosting a protease (red) that catalyzes the breakage of the substrate into smaller peptide fragments. Right: Prof. Dr. Barbara Saccà.
Further Information:
https://www.nature.com/articles/s41565-024-01738-7
Prof. Dr. Barbara Sacca, Faculty of Biology and Center of Medical Biotechnology, Tel. 0201/18 3-3395, barbara.sacca@uni-due.de
Editorial office: Jennifer Meina, Tel. 0203/379-1205, jennifer.meina@uni-due.de
