Research
Current Directions

Research overview (All images: © B.Saccà)
Financial Support
- SFB 1093
- DFG
- Mercator Stiftung
- Deutsche Krebshilfe
Spatial Confinement
Our DNA-origami nanocontainers rely on a host-guest matching principle to bind specific proteins. We aim to understand how spatial confinement can affect the thermodynamics and kinetics of molecular interactions under various conditions with an emphasis on their potential for DNA-based information processing.
Hierarchical Assembly
Natural macromolecular assemblies achieve complexity through a hierarchical paradigm that relies on step-wise interactions between shape-complementary subunits. We apply this design principle to create DNA assemblies with specific structural properties.
Dynamic Response
Biological systems can sense and respond to external events through structural reconfiguration or activation of distinct paths. This enables to control critical functions and carry out various tasks as required. We use switchable DNA motifs, DNA logic gates, and DNA reaction cascades integrated within static DNA nanostructures to mimic this behavior.
- Engineering modular enzymes using DNA origami
- Sequence-dependent folding of DNA origami
- A switchable five-states device
- Growth and stability of DNA origami filaments
- The effect of DNA-confinement on the proteolytic activity of thrombin
- Travelling the energy landscape of DNA origami folding
- DNA filaments with different ultrastructures and persistence lengths
- A spring-like DNA origami device
- Rational design of DNA hosts for protein guests
- Hierarchical self-assembly of DNA nanostructures
Engineering modular enzymes using DNA origami
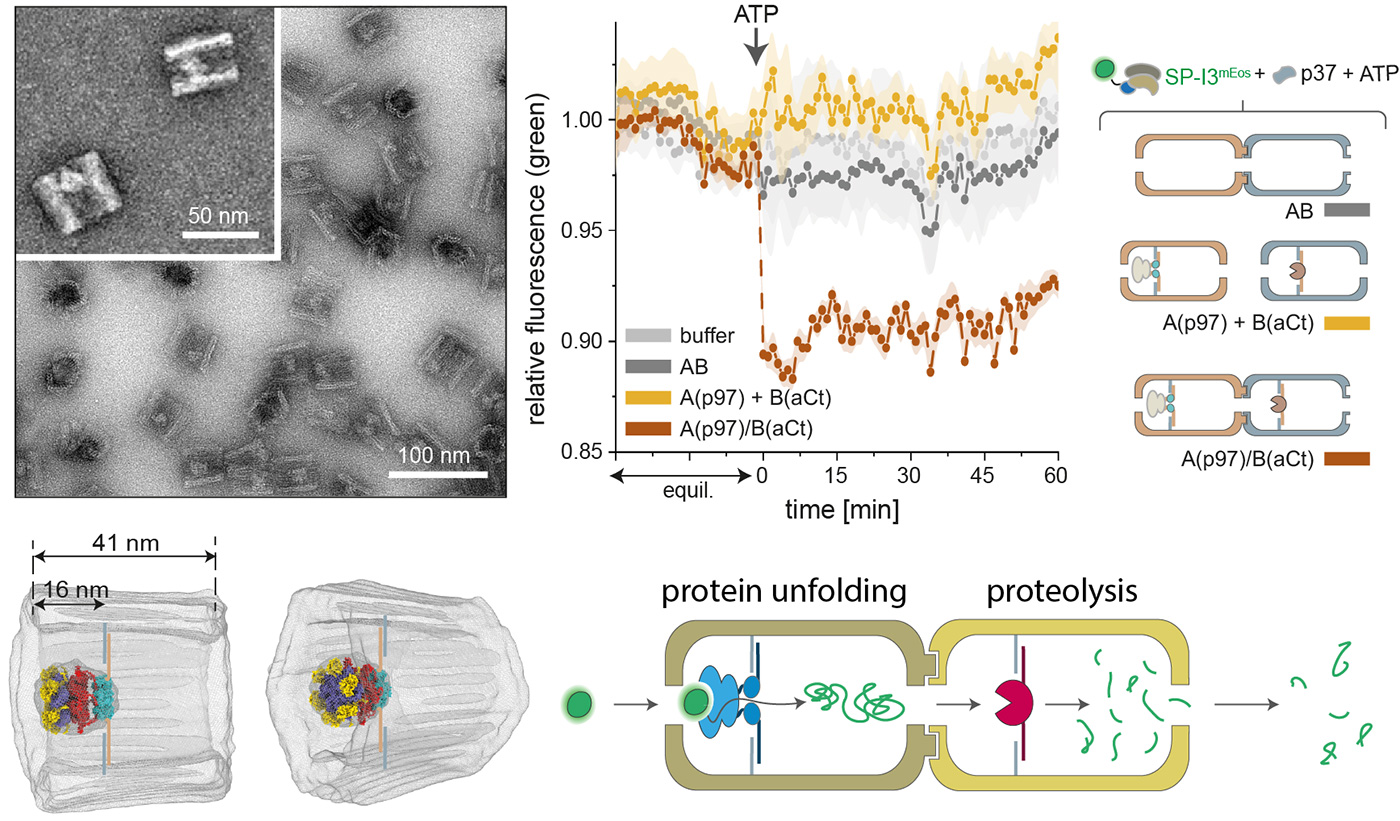
A DNA origami nanocompartment is designed to trap an unfoldase machine in a unidirectional orientation. This provides a gateway mechanism for substrate recruitment and translocation to a downstream compartment, hosting a protease. Kinetics and proteomics data demonstrate that the physical connection of the DNA-based modules improves the global performance of the chimera and reduces off-target reactions.
Sequence-dependent folding of DNA origami
Current DNA origami folding models provide insight into the yield of the assembly process and the structural isomerization under mechanical forces. In this study, we investigate the impact of sequence on the folding of small DNA origami domains into one of two possible isomers and propose thermal criton maps to interpret our findings.
A switchable five-states device
In this work, we constructed a reconfigurable nanodevice comprised of two DNA boxes in a face-to-face orientation mounted on top of a DNA platform. Addition of fuel/antifuel strands drives the reversible switch of the platform among an OFF state and one of five possible ON states, which differ in the distance between the two halves of the platform (from 0 bp to 43 bp).
Growth and stability of DNA origami filaments
DNA origami filaments are of great interest due to their versatile morphologies, mechanical properties, and functionalities. In this study, we explore the thermodynamic and kinetic properties of linear structures formed from a ditopic DNA origami monomer with two distinct interfaces. We investigate how base-hybridization or base-stacking interactions can trigger the dimerization and polymerization process and demonstrate how hierarchical DNA origami assembly can be influenced by the anisotropy of building units and their self-association modes.
The effect of DNA-confinement on the proteolytic activity of thrombin
In this study, we employed thrombin as a model for allosterically regulated serine proteases, and used aptamers to confine the enzyme within various types of DNA origami cavities, differing in their structural and electrostatic features. By comparing the hydrolysis of substrates with distinct net charge, we found that the reaction rate is influenced by the electrostatic interactions between the DNA cage and the substrate, and depends on the extent of tethering between the cage and the enzyme. For substrates with opposite net charge, this results in a reversal of the catalytic response of DNA-scaffolded thrombin compared to freely diffusing thrombin. Spatial confinement within DNA compartments can be therefore used as a tool for regulating allosteric processes.
Travelling the energy landscape of DNA origami folding
The self-assembly of a DNA origami structure, although mostly feasible, represents indeed a rather complex folding problem. Entropy-driven folding and nucleation seeds formation may provide possible solutions; however, until now, a unified view of the energetic factors in play is missing. By analyzing the self-assembly of origami domains with identical structure but different nucleobase composition, in function of variable design and experimental parameters, we identify the role played by sequence-dependent forces at the edges of the structure, where topological constraint is higher. Our data show that the degree of mechanical stress experienced by these regions during initial folding reshapes the energy landscape profile, defining the ratio between two possible global conformations. We thus propose a dynamic model of DNA origami assembly that relies on the capability of the system to escape high structural frustration at nucleation sites, eventually resulting in the emergence of a more favorable but previously hidden state.
DNA filaments with different ultrastructures and persistence lengths
Using DNA origami approaches, we constructed a reconfigurable module, composed of two quasi-independent domains and four possible interfaces, capable of facial and lateral growing through specific recognition patterns. Whereas the flexibility of the intra-domains region can be regulated by switchable DNA motifs, the inter-domain interfaces feature mutually and self-complementary shapes, whose pairwise association leads to filaments of programmable periodicity and variable persistence length. In this work, we show that the assembly pathway leading to oligomeric chains can be finely tuned and fully controlled, enabling to emulate the formation of protein-like filaments using a single construction principle. This leads to artificial materials with a large variety of ultrastructures and bending strengths comparable, or even superior, to their natural counterparts.
A spring-like DNA origami device
In this work, we used temperature-dependent FRET spectroscopy to extract the thermal stabilities of distinct sets of hairpins integrated into the central seam of a DNA origami structure. We developed a hybrid spring model to describe the energy landscape of the tethered hairpins, combining the thermodynamic nearest-neighbor energy of duplex DNA with the entropic free energy of single-stranded DNA estimated using a worm-like chain approximation. We show that the organized scaffolding of multiple hairpins enhances the thermal stability of the device and that the coordinated action of the tethered motors can be used to mechanically unfold a G-quadruplex motif bound into the inner cavity of the origami structure, thus surpassing the operational capabilities of freely diffusing motors.
Rational design of DNA hosts for protein guests
Natural host-guest complexes are normally stabilized by non-covalent interactions between geometrically matching surfaces. Using this principle, we realized one of the largest DNA-protein complexes of semisynthetic origin held in place exclusively by spatially defined supramolecular interactions. Our approach relies on the decoration of the inner surface of a hollow structure with multiple ligands converging to their corresponding binding sites on the protein surface with programmable symmetry and range-of-action.

A. Sprengel, P. Lill, P. Stegemann, K. Bravo-Rodriguez, E.-C. Schöneweiß, M. Merdanovic, D. Gudnason, M. Aznauryan, L. Gamrad, S. Barcikowski, E. Sanchez-Garcia, V. Birkedal, C. Gatsogiannis, M. Ehrmann, B. Saccà. Tailored protein encapsulation into a DNA host using geometrically organized supramolecular interactions. Nat. Commun. 2017, 8, 14472.
Hierarchical self-assembly of DNA nanostructures
From atoms to molecules and biomacromolecules, from organelles to cells, tissues, till the whole living system: so nature shows us that the realization of complex systems with emergent properties originates from the hierarchical self-assembly of single components in a guided bottom-up process. Using DNA as a fundamental building block with well-known self-recognition properties, scientists have developed design rules and physical-chemical approaches for the fully programmable construction of highly organized structures with nanosized features.